9.5 Issues with GLM (and regression analyses in general)
9.5.1 Correlation tests
Below, we look at two predictors that are correlated with each other: Z3-Z2 (F3-F2 in Bark) and A2*-A3* (normalised amplitude differences between harmonics closest to F2 and F3). The results of the correlation test shows the two predictors to negatively correlate with each other at a rate of -0.87.
##
## Pearson's product-moment correlation
##
## data: dfPharV2$Z3mnZ2 and dfPharV2$A2mnA3
## t = -35.864, df = 400, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8947506 -0.8480066
## sample estimates:
## cor
## -0.87337519.5.2 Plots to visualise the data
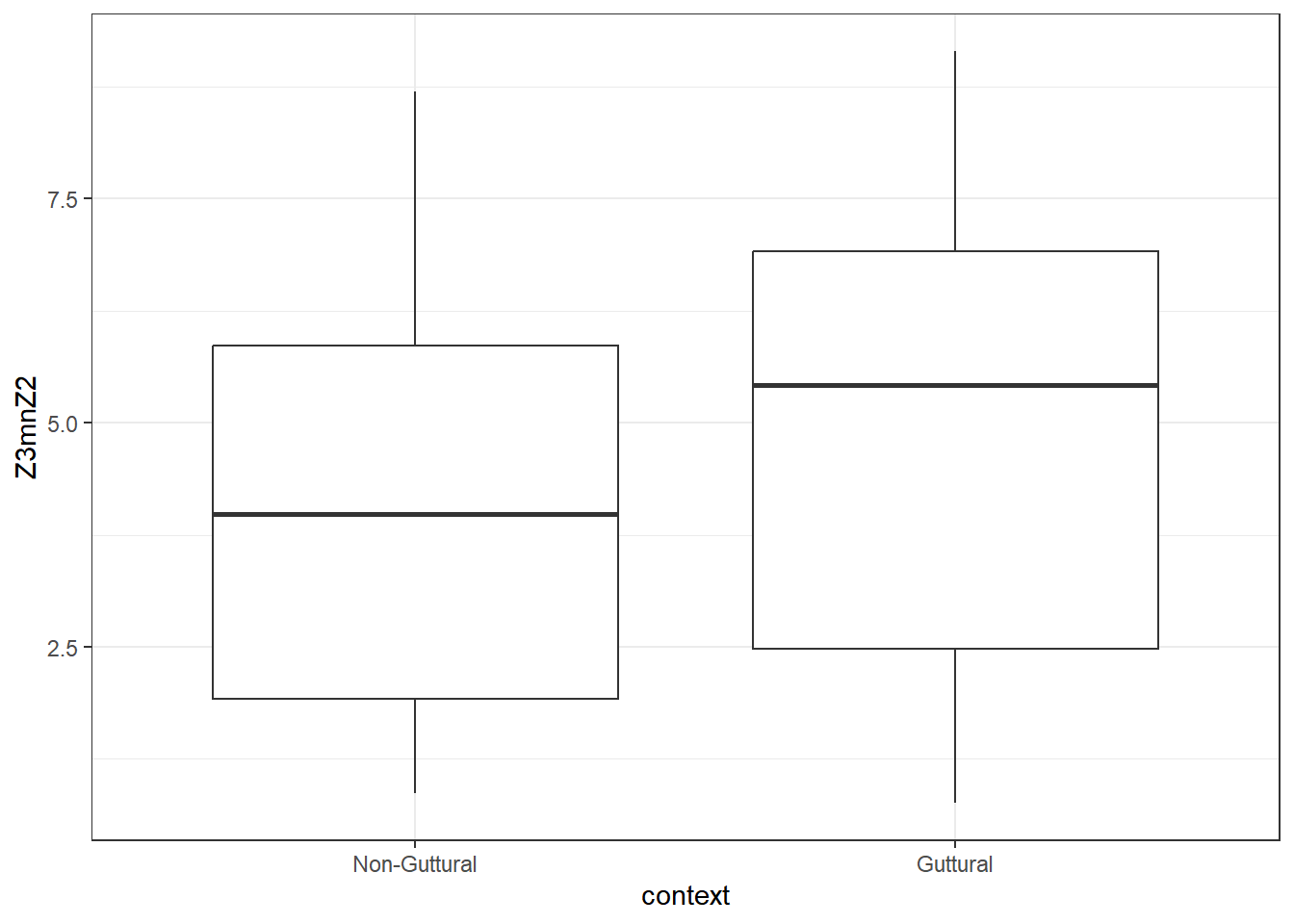
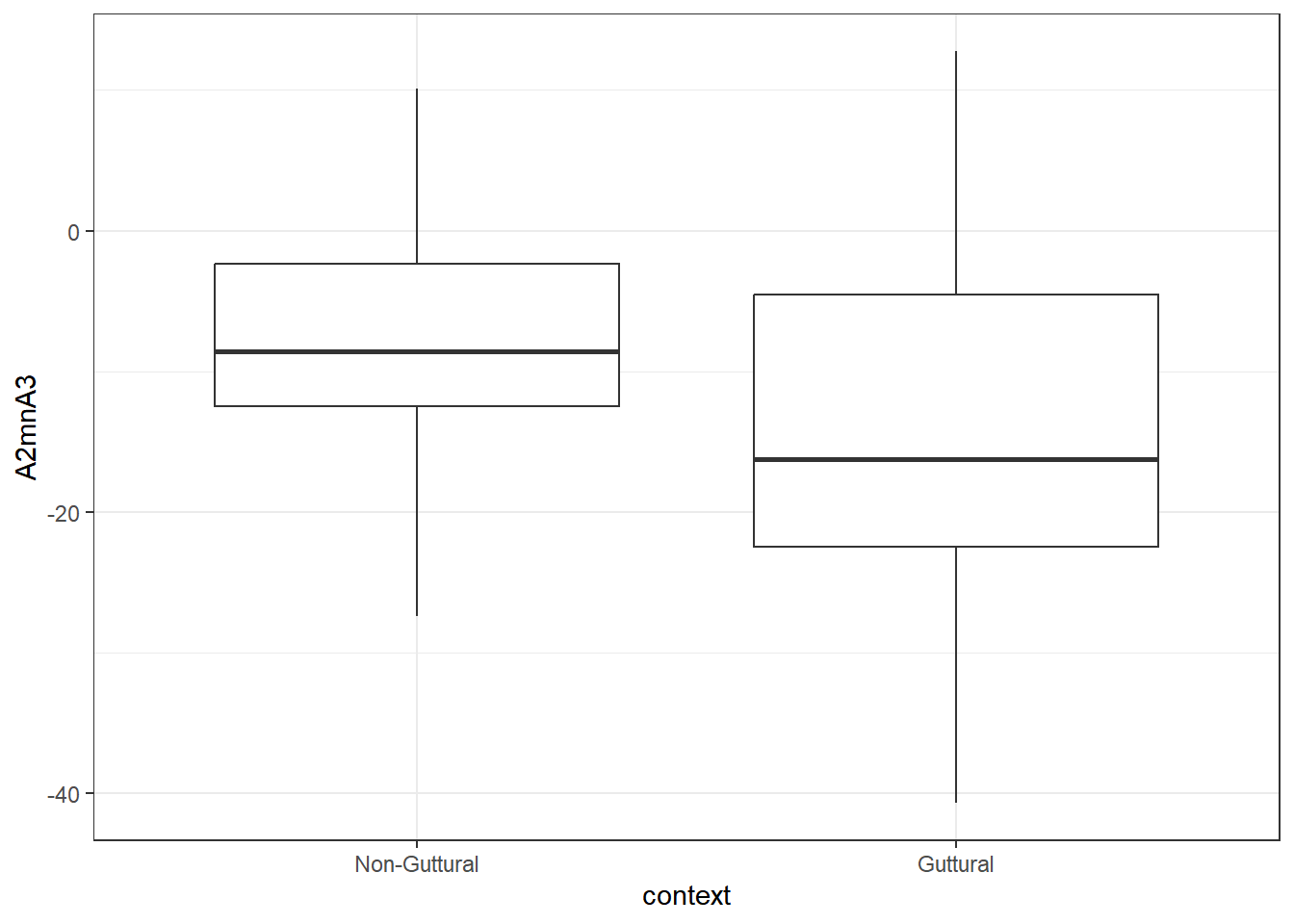
As we see from the plot, Z3-Z2 is higher in the guttural, whereas A2*-A3* is lower.
9.5.4 Conclusion
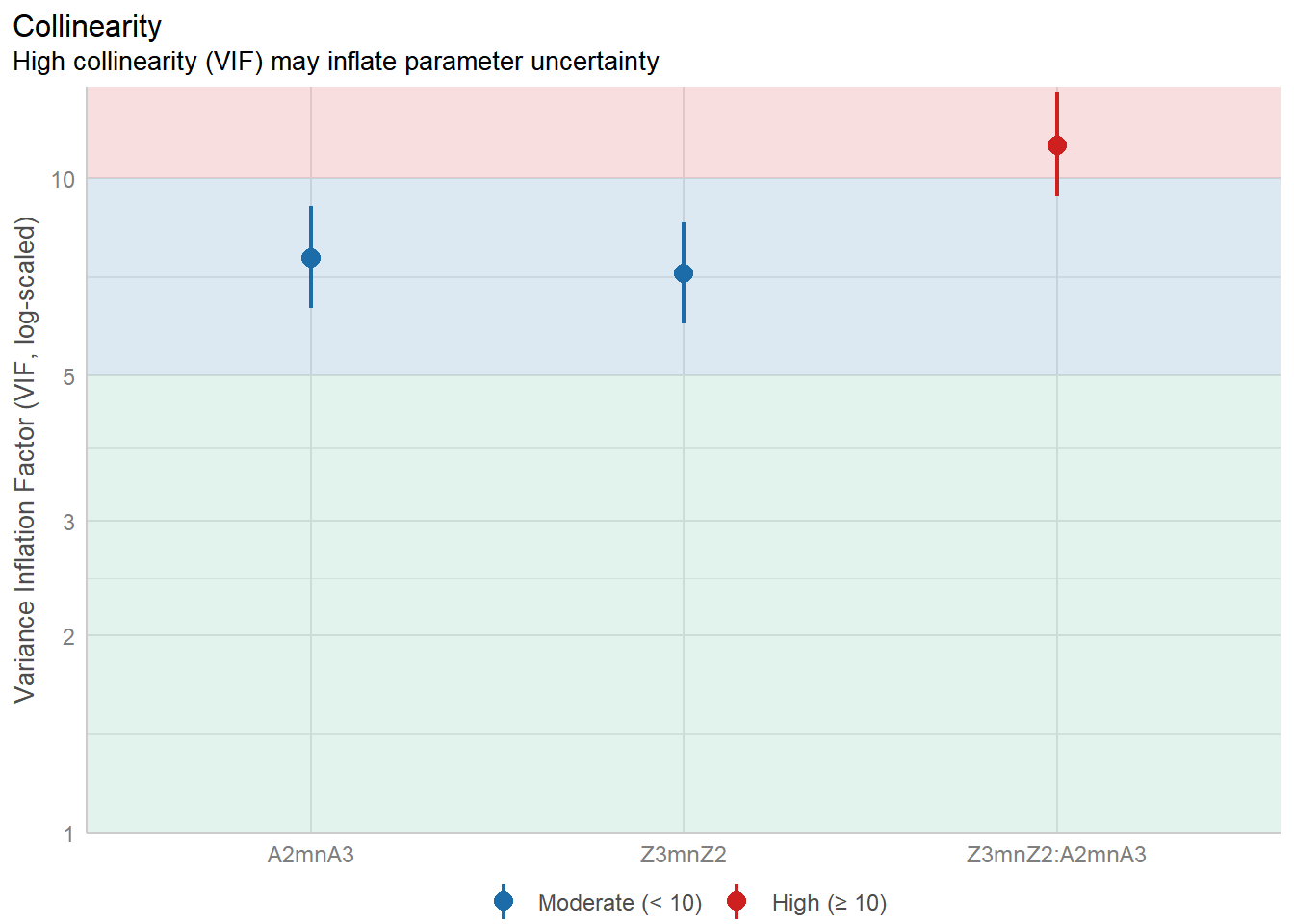
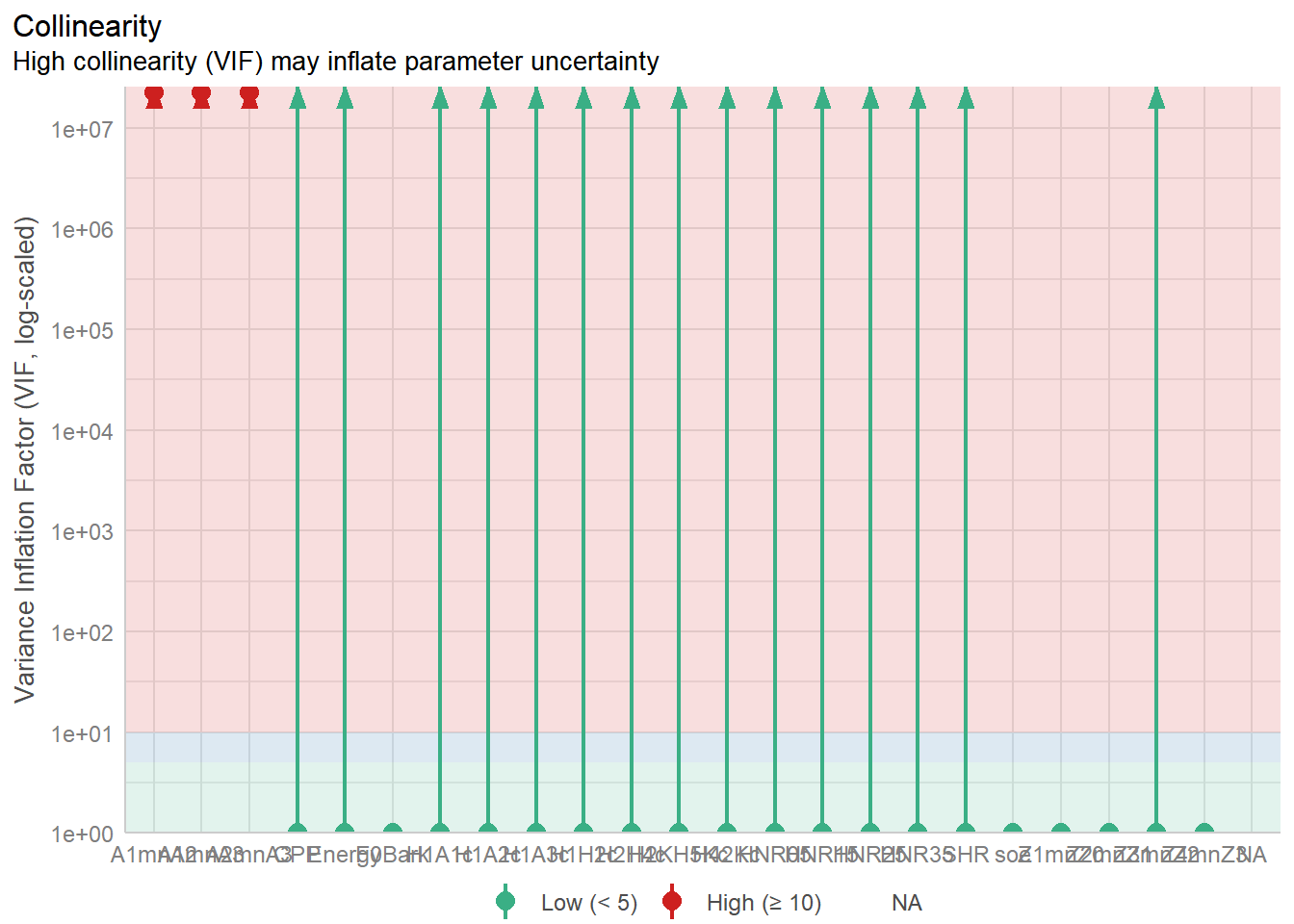
The more complex your regression model becomes, the higher level of collinearity one can see. This is a known issue in regression analyses, and without applying appropriate approaches, e.g., verifying the collinearity in the data; choosing non-correlated predictors, etc.., the models become inflated and the confidence in the results becomes reduced.
There are various solutions to this issue, such as removing collinear predictors, using PCA to obtain orthogonal dimension (i.e., non-correlated dimensions), etc.. Another solution is to use decision trees and Random Forest. Both are very well known to deal with collinearity and to make sense of multivariate predictors.