10.5 Issues with GLM (and regression analyses in general)
10.5.1 Correlation tests
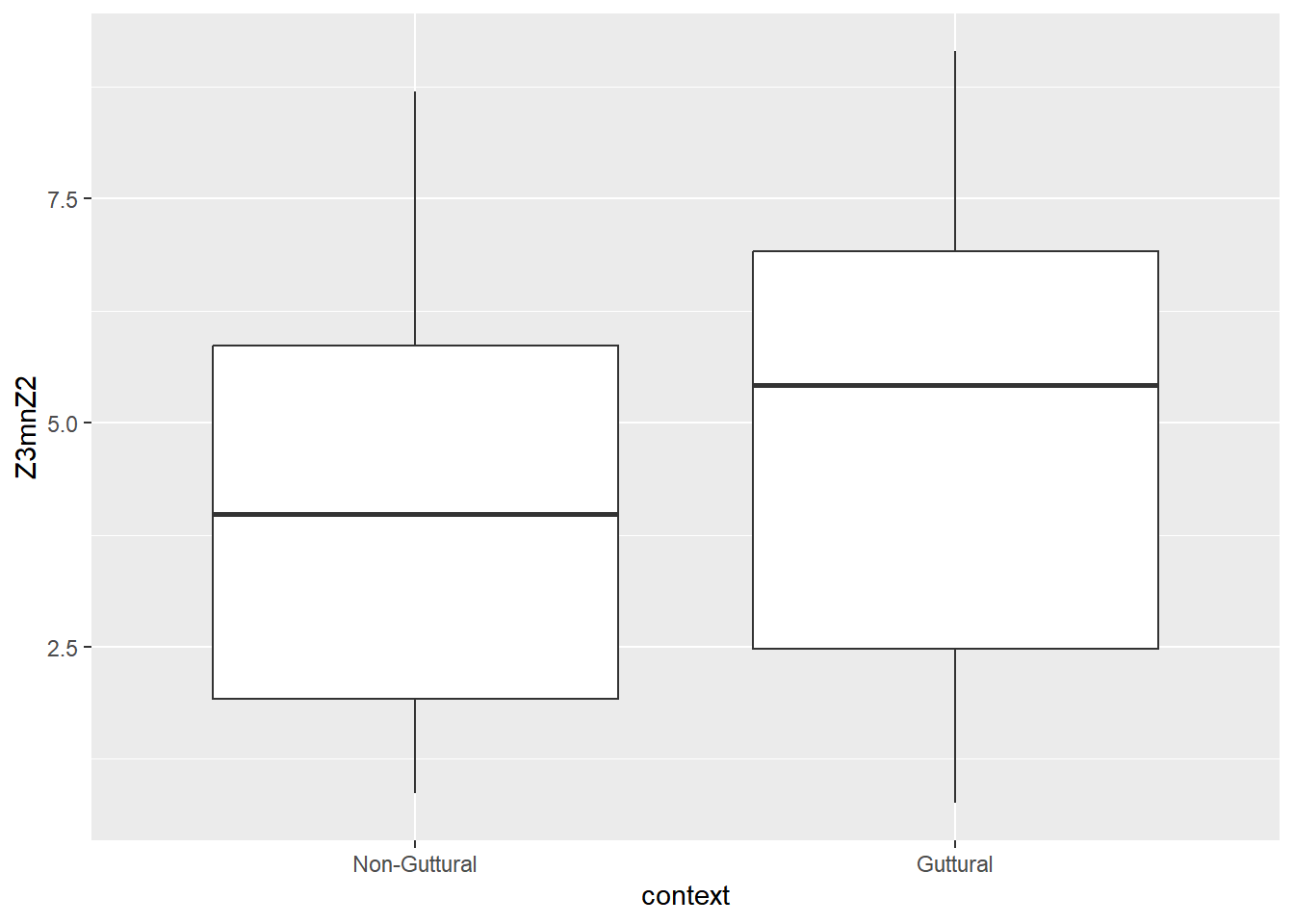
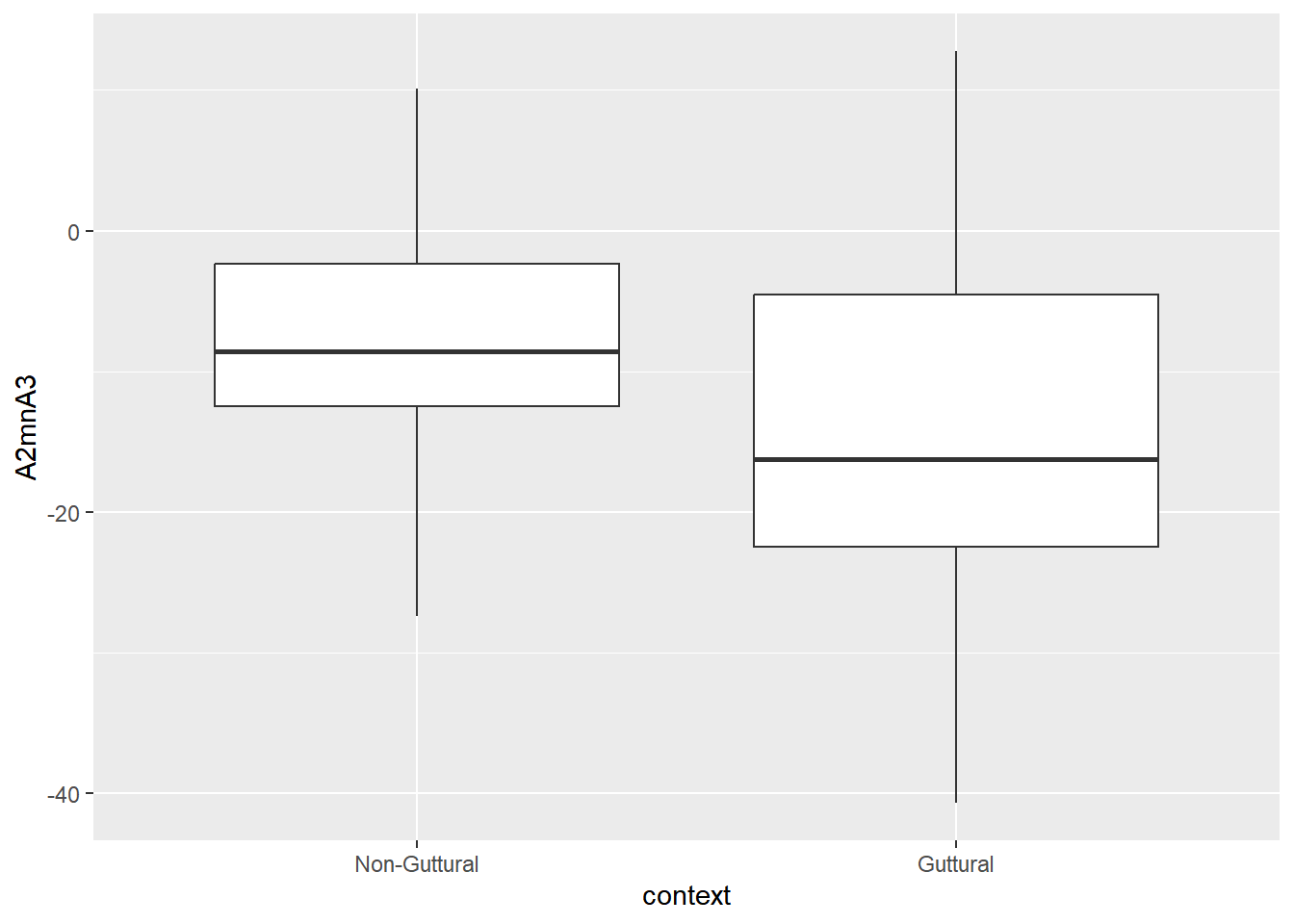
Below, we look at two predictors that are correlated with each other: Z3-Z2 (F3-F2 in Bark) and A2*-A3* (normalised amplitude differences between harmonics closest to F2 and F3). The results of the correlation test shows the two predictors to negatively correlate with each other at a rate of -0.87.
##
## Pearson's product-moment correlation
##
## data: dfPharV2$Z3mnZ2 and dfPharV2$A2mnA3
## t = -35.864, df = 400, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.8947506 -0.8480066
## sample estimates:
## cor
## -0.8733751