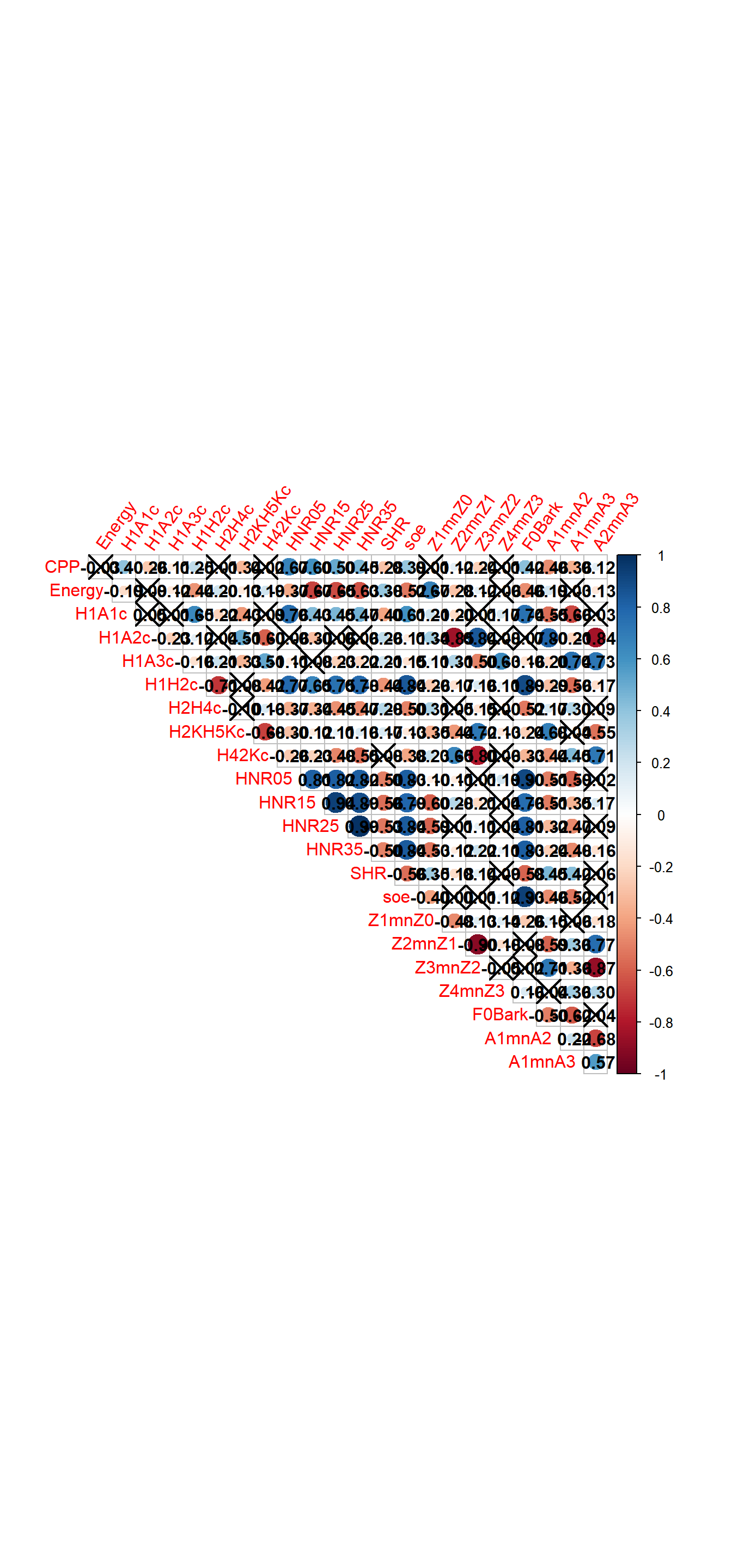
9.3 Correlation tests
Above, we did a correlation test on two predictors.
What if we want to obtain a nice plot of all numeric predictors and add significance levels? We use the package corrplot
9.3.1 Correlation plots
9.3.1.1 Correlation plots 1
## CPP Energy H1A1c H1A2c H1A3c
## CPP 1.000000000 -0.03399038 0.404706765 -0.26455286 -0.113482532
## Energy -0.033990377 1.00000000 -0.186476670 0.09170318 -0.122377040
## H1A1c 0.404706765 -0.18647667 1.000000000 0.05465872 0.007167585
## H1A2c -0.264552856 0.09170318 0.054658717 1.00000000 -0.233216979
## H1A3c -0.113482532 -0.12237704 0.007167585 -0.23321698 1.000000000
## H1H2c 0.251810189 -0.44444137 0.648990000 0.11615684 -0.164917390
## H2H4c -0.005458994 0.20371885 -0.220955156 0.04292950 0.210063534
## H2KH5Kc -0.336168212 -0.12504346 -0.431991841 0.51057160 -0.334656923
## H42Kc 0.019225753 0.19493845 -0.090631471 -0.59694938 0.514544301
## HNR05 0.666184301 -0.37052021 0.758327365 -0.06157287 -0.108056123
## HNR15 0.603409742 -0.67352331 0.433216471 -0.30096436 -0.075490785
## HNR25 0.504781686 -0.65091028 0.451043577 -0.06083878 -0.231125560
## HNR35 0.453677497 -0.63296467 0.470614004 0.05539693 -0.216734533
## SHR -0.284829618 0.38017553 -0.395846797 0.25865972 0.209623680
## soe 0.386759609 -0.52487995 0.609437744 -0.11463910 -0.153217238
## Z1mnZ0 0.014248500 0.67114927 0.213674458 0.33815761 0.109502228
## Z2mnZ1 0.121821865 -0.28394211 -0.197144132 -0.84586885 0.307462932
## Z3mnZ2 -0.236018438 0.11604624 -0.014226757 0.84163102 -0.496911115
## Z4mnZ3 -0.009359468 -0.06257675 0.173172293 0.07552205 0.630948791
## F0Bark 0.417866035 -0.46400144 0.742753093 -0.06972759 -0.162186194
## A1mnA2 -0.462940266 0.18810560 -0.552669620 0.80194637 -0.198942082
## A1mnA3 -0.355892004 0.03344952 -0.664469548 -0.21090543 0.742533502
## A2mnA3 0.122501972 -0.13362699 -0.034489484 -0.83635261 0.728126945
## H1H2c H2H4c H2KH5Kc H42Kc HNR05
## CPP 0.25181019 -0.005458994 -0.33616821 0.01922575 0.66618430
## Energy -0.44444137 0.203718846 -0.12504346 0.19493845 -0.37052021
## H1A1c 0.64899000 -0.220955156 -0.43199184 -0.09063147 0.75832736
## H1A2c 0.11615684 0.042929498 0.51057160 -0.59694938 -0.06157287
## H1A3c -0.16491739 0.210063534 -0.33465692 0.51454430 -0.10805612
## H1H2c 1.00000000 -0.713751116 -0.07796057 -0.41916885 0.77141984
## H2H4c -0.71375112 1.000000000 -0.09596636 0.16166922 -0.36847378
## H2KH5Kc -0.07796057 -0.095966364 1.00000000 -0.68312931 -0.29753363
## H42Kc -0.41916885 0.161669220 -0.68312931 1.00000000 -0.26285288
## HNR05 0.77141984 -0.368473783 -0.29753363 -0.26285288 1.00000000
## HNR15 0.65474601 -0.344119338 -0.11535020 -0.22732173 0.81480902
## HNR25 0.74616221 -0.451131758 0.10665010 -0.48042235 0.82316561
## HNR35 0.77808181 -0.471273417 0.15703813 -0.55230135 0.82336788
## SHR -0.44306719 0.283034424 0.16958475 0.07533203 -0.50080260
## soe 0.84462374 -0.497528429 -0.12742022 -0.38269875 0.82808974
## Z1mnZ0 -0.25783681 0.314095793 -0.35109828 0.22859438 -0.10404198
## Z2mnZ1 -0.16754562 0.052215339 -0.43864642 0.65068250 -0.10884383
## Z3mnZ2 0.16165571 -0.150529963 0.71841244 -0.80426718 -0.01280049
## Z4mnZ3 0.10977975 0.003456879 -0.13027105 0.06309325 0.19394214
## F0Bark 0.89121654 -0.522808166 -0.23993155 -0.33335794 0.90374524
## A1mnA2 -0.29133499 0.168027465 0.68460673 -0.44401978 -0.50509144
## A1mnA3 -0.55795935 0.304989227 0.03925946 0.44524464 -0.58870128
## A2mnA3 -0.17484791 0.088160327 -0.54854966 0.71084573 -0.01751585
## HNR15 HNR25 HNR35 SHR soe
## CPP 0.60340974 0.504781686 0.45367750 -0.28482962 0.386759609
## Energy -0.67352331 -0.650910279 -0.63296467 0.38017553 -0.524879953
## H1A1c 0.43321647 0.451043577 0.47061400 -0.39584680 0.609437744
## H1A2c -0.30096436 -0.060838775 0.05539693 0.25865972 -0.114639103
## H1A3c -0.07549078 -0.231125560 -0.21673453 0.20962368 -0.153217238
## H1H2c 0.65474601 0.746162213 0.77808181 -0.44306719 0.844623735
## H2H4c -0.34411934 -0.451131758 -0.47127342 0.28303442 -0.497528429
## H2KH5Kc -0.11535020 0.106650103 0.15703813 0.16958475 -0.127420222
## H42Kc -0.22732173 -0.480422351 -0.55230135 0.07533203 -0.382698752
## HNR05 0.81480902 0.823165610 0.82336788 -0.50080260 0.828089735
## HNR15 1.00000000 0.938770768 0.88727082 -0.55580661 0.792886846
## HNR25 0.93877077 1.000000000 0.98644454 -0.52810263 0.837725385
## HNR35 0.88727082 0.986444536 1.00000000 -0.49891921 0.839675263
## SHR -0.55580661 -0.528102632 -0.49891921 1.00000000 -0.555384532
## soe 0.79288685 0.837725385 0.83967526 -0.55538453 1.000000000
## Z1mnZ0 -0.59593537 -0.589202134 -0.53287118 0.35389648 -0.403168468
## Z2mnZ1 0.28433812 0.009867765 -0.11639529 -0.18153788 -0.004677426
## Z3mnZ2 -0.20572721 0.109264859 0.21831675 0.13689371 0.007849427
## Z4mnZ3 0.04324078 0.035922107 0.11146279 0.08718854 0.117966357
## F0Bark 0.76082881 0.812555958 0.82810219 -0.58077659 0.934247247
## A1mnA2 -0.51038750 -0.320632658 -0.23532567 0.45272098 -0.460304226
## A1mnA3 -0.34659555 -0.474847635 -0.47720189 0.42180815 -0.522721261
## A2mnA3 0.16958474 -0.087411661 -0.16123104 -0.06414896 -0.005568759
## Z1mnZ0 Z2mnZ1 Z3mnZ2 Z4mnZ3 F0Bark
## CPP 0.01424850 0.121821865 -0.236018438 -0.009359468 0.41786604
## Energy 0.67114927 -0.283942110 0.116046236 -0.062576748 -0.46400144
## H1A1c 0.21367446 -0.197144132 -0.014226757 0.173172293 0.74275309
## H1A2c 0.33815761 -0.845868849 0.841631017 0.075522047 -0.06972759
## H1A3c 0.10950223 0.307462932 -0.496911115 0.630948791 -0.16218619
## H1H2c -0.25783681 -0.167545617 0.161655712 0.109779752 0.89121654
## H2H4c 0.31409579 0.052215339 -0.150529963 0.003456879 -0.52280817
## H2KH5Kc -0.35109828 -0.438646418 0.718412444 -0.130271054 -0.23993155
## H42Kc 0.22859438 0.650682503 -0.804267177 0.063093254 -0.33335794
## HNR05 -0.10404198 -0.108843828 -0.012800492 0.193942142 0.90374524
## HNR15 -0.59593537 0.284338125 -0.205727207 0.043240778 0.76082881
## HNR25 -0.58920213 0.009867765 0.109264859 0.035922107 0.81255596
## HNR35 -0.53287118 -0.116395288 0.218316751 0.111462787 0.82810219
## SHR 0.35389648 -0.181537883 0.136893713 0.087188543 -0.58077659
## soe -0.40316847 -0.004677426 0.007849427 0.117966357 0.93424725
## Z1mnZ0 1.00000000 -0.479471463 0.132402681 0.143899368 -0.25634372
## Z2mnZ1 -0.47947146 1.000000000 -0.901393170 -0.176529973 -0.08020731
## Z3mnZ2 0.13240268 -0.901393170 1.000000000 -0.050618065 0.01898548
## Z4mnZ3 0.14389937 -0.176529973 -0.050618065 1.000000000 0.15981826
## F0Bark -0.25634372 -0.080207311 0.018985484 0.159818261 1.00000000
## A1mnA2 0.15440193 -0.588053094 0.710977937 -0.040573920 -0.50258069
## A1mnA3 -0.06129172 0.361831277 -0.361830045 0.355533137 -0.61872264
## A2mnA3 -0.17662983 0.769563482 -0.873375083 0.302455298 -0.04228227
## A1mnA2 A1mnA3 A2mnA3
## CPP -0.46294027 -0.35589200 0.122501972
## Energy 0.18810560 0.03344952 -0.133626988
## H1A1c -0.55266962 -0.66446955 -0.034489484
## H1A2c 0.80194637 -0.21090543 -0.836352608
## H1A3c -0.19894208 0.74253350 0.728126945
## H1H2c -0.29133499 -0.55795935 -0.174847909
## H2H4c 0.16802747 0.30498923 0.088160327
## H2KH5Kc 0.68460673 0.03925946 -0.548549664
## H42Kc -0.44401978 0.44524464 0.710845732
## HNR05 -0.50509144 -0.58870128 -0.017515845
## HNR15 -0.51038750 -0.34659555 0.169584736
## HNR25 -0.32063266 -0.47484763 -0.087411661
## HNR35 -0.23532567 -0.47720189 -0.161231042
## SHR 0.45272098 0.42180815 -0.064148964
## soe -0.46030423 -0.52272126 -0.005568759
## Z1mnZ0 0.15440193 -0.06129172 -0.176629833
## Z2mnZ1 -0.58805309 0.36183128 0.769563482
## Z3mnZ2 0.71097794 -0.36183005 -0.873375083
## Z4mnZ3 -0.04057392 0.35553314 0.302455298
## F0Bark -0.50258069 -0.61872264 -0.042282270
## A1mnA2 1.00000000 0.22151418 -0.677425247
## A1mnA3 0.22151418 1.00000000 0.567258129
## A2mnA3 -0.67742525 0.56725813 1.000000000## CPP Energy H1A1c H1A2c H1A3c
## CPP 1.000000000 -0.03399038 0.404706765 -0.26455286 -0.113482532
## Energy -0.033990377 1.00000000 -0.186476670 0.09170318 -0.122377040
## H1A1c 0.404706765 -0.18647667 1.000000000 0.05465872 0.007167585
## H1A2c -0.264552856 0.09170318 0.054658717 1.00000000 -0.233216979
## H1A3c -0.113482532 -0.12237704 0.007167585 -0.23321698 1.000000000
## H1H2c 0.251810189 -0.44444137 0.648990000 0.11615684 -0.164917390
## H2H4c -0.005458994 0.20371885 -0.220955156 0.04292950 0.210063534
## H2KH5Kc -0.336168212 -0.12504346 -0.431991841 0.51057160 -0.334656923
## H42Kc 0.019225753 0.19493845 -0.090631471 -0.59694938 0.514544301
## HNR05 0.666184301 -0.37052021 0.758327365 -0.06157287 -0.108056123
## HNR15 0.603409742 -0.67352331 0.433216471 -0.30096436 -0.075490785
## HNR25 0.504781686 -0.65091028 0.451043577 -0.06083878 -0.231125560
## HNR35 0.453677497 -0.63296467 0.470614004 0.05539693 -0.216734533
## SHR -0.284829618 0.38017553 -0.395846797 0.25865972 0.209623680
## soe 0.386759609 -0.52487995 0.609437744 -0.11463910 -0.153217238
## Z1mnZ0 0.014248500 0.67114927 0.213674458 0.33815761 0.109502228
## Z2mnZ1 0.121821865 -0.28394211 -0.197144132 -0.84586885 0.307462932
## Z3mnZ2 -0.236018438 0.11604624 -0.014226757 0.84163102 -0.496911115
## Z4mnZ3 -0.009359468 -0.06257675 0.173172293 0.07552205 0.630948791
## F0Bark 0.417866035 -0.46400144 0.742753093 -0.06972759 -0.162186194
## A1mnA2 -0.462940266 0.18810560 -0.552669620 0.80194637 -0.198942082
## A1mnA3 -0.355892004 0.03344952 -0.664469548 -0.21090543 0.742533502
## A2mnA3 0.122501972 -0.13362699 -0.034489484 -0.83635261 0.728126945
## H1H2c H2H4c H2KH5Kc H42Kc HNR05
## CPP 0.25181019 -0.005458994 -0.33616821 0.01922575 0.66618430
## Energy -0.44444137 0.203718846 -0.12504346 0.19493845 -0.37052021
## H1A1c 0.64899000 -0.220955156 -0.43199184 -0.09063147 0.75832736
## H1A2c 0.11615684 0.042929498 0.51057160 -0.59694938 -0.06157287
## H1A3c -0.16491739 0.210063534 -0.33465692 0.51454430 -0.10805612
## H1H2c 1.00000000 -0.713751116 -0.07796057 -0.41916885 0.77141984
## H2H4c -0.71375112 1.000000000 -0.09596636 0.16166922 -0.36847378
## H2KH5Kc -0.07796057 -0.095966364 1.00000000 -0.68312931 -0.29753363
## H42Kc -0.41916885 0.161669220 -0.68312931 1.00000000 -0.26285288
## HNR05 0.77141984 -0.368473783 -0.29753363 -0.26285288 1.00000000
## HNR15 0.65474601 -0.344119338 -0.11535020 -0.22732173 0.81480902
## HNR25 0.74616221 -0.451131758 0.10665010 -0.48042235 0.82316561
## HNR35 0.77808181 -0.471273417 0.15703813 -0.55230135 0.82336788
## SHR -0.44306719 0.283034424 0.16958475 0.07533203 -0.50080260
## soe 0.84462374 -0.497528429 -0.12742022 -0.38269875 0.82808974
## Z1mnZ0 -0.25783681 0.314095793 -0.35109828 0.22859438 -0.10404198
## Z2mnZ1 -0.16754562 0.052215339 -0.43864642 0.65068250 -0.10884383
## Z3mnZ2 0.16165571 -0.150529963 0.71841244 -0.80426718 -0.01280049
## Z4mnZ3 0.10977975 0.003456879 -0.13027105 0.06309325 0.19394214
## F0Bark 0.89121654 -0.522808166 -0.23993155 -0.33335794 0.90374524
## A1mnA2 -0.29133499 0.168027465 0.68460673 -0.44401978 -0.50509144
## A1mnA3 -0.55795935 0.304989227 0.03925946 0.44524464 -0.58870128
## A2mnA3 -0.17484791 0.088160327 -0.54854966 0.71084573 -0.01751585
## HNR15 HNR25 HNR35 SHR soe
## CPP 0.60340974 0.504781686 0.45367750 -0.28482962 0.386759609
## Energy -0.67352331 -0.650910279 -0.63296467 0.38017553 -0.524879953
## H1A1c 0.43321647 0.451043577 0.47061400 -0.39584680 0.609437744
## H1A2c -0.30096436 -0.060838775 0.05539693 0.25865972 -0.114639103
## H1A3c -0.07549078 -0.231125560 -0.21673453 0.20962368 -0.153217238
## H1H2c 0.65474601 0.746162213 0.77808181 -0.44306719 0.844623735
## H2H4c -0.34411934 -0.451131758 -0.47127342 0.28303442 -0.497528429
## H2KH5Kc -0.11535020 0.106650103 0.15703813 0.16958475 -0.127420222
## H42Kc -0.22732173 -0.480422351 -0.55230135 0.07533203 -0.382698752
## HNR05 0.81480902 0.823165610 0.82336788 -0.50080260 0.828089735
## HNR15 1.00000000 0.938770768 0.88727082 -0.55580661 0.792886846
## HNR25 0.93877077 1.000000000 0.98644454 -0.52810263 0.837725385
## HNR35 0.88727082 0.986444536 1.00000000 -0.49891921 0.839675263
## SHR -0.55580661 -0.528102632 -0.49891921 1.00000000 -0.555384532
## soe 0.79288685 0.837725385 0.83967526 -0.55538453 1.000000000
## Z1mnZ0 -0.59593537 -0.589202134 -0.53287118 0.35389648 -0.403168468
## Z2mnZ1 0.28433812 0.009867765 -0.11639529 -0.18153788 -0.004677426
## Z3mnZ2 -0.20572721 0.109264859 0.21831675 0.13689371 0.007849427
## Z4mnZ3 0.04324078 0.035922107 0.11146279 0.08718854 0.117966357
## F0Bark 0.76082881 0.812555958 0.82810219 -0.58077659 0.934247247
## A1mnA2 -0.51038750 -0.320632658 -0.23532567 0.45272098 -0.460304226
## A1mnA3 -0.34659555 -0.474847635 -0.47720189 0.42180815 -0.522721261
## A2mnA3 0.16958474 -0.087411661 -0.16123104 -0.06414896 -0.005568759
## Z1mnZ0 Z2mnZ1 Z3mnZ2 Z4mnZ3 F0Bark
## CPP 0.01424850 0.121821865 -0.236018438 -0.009359468 0.41786604
## Energy 0.67114927 -0.283942110 0.116046236 -0.062576748 -0.46400144
## H1A1c 0.21367446 -0.197144132 -0.014226757 0.173172293 0.74275309
## H1A2c 0.33815761 -0.845868849 0.841631017 0.075522047 -0.06972759
## H1A3c 0.10950223 0.307462932 -0.496911115 0.630948791 -0.16218619
## H1H2c -0.25783681 -0.167545617 0.161655712 0.109779752 0.89121654
## H2H4c 0.31409579 0.052215339 -0.150529963 0.003456879 -0.52280817
## H2KH5Kc -0.35109828 -0.438646418 0.718412444 -0.130271054 -0.23993155
## H42Kc 0.22859438 0.650682503 -0.804267177 0.063093254 -0.33335794
## HNR05 -0.10404198 -0.108843828 -0.012800492 0.193942142 0.90374524
## HNR15 -0.59593537 0.284338125 -0.205727207 0.043240778 0.76082881
## HNR25 -0.58920213 0.009867765 0.109264859 0.035922107 0.81255596
## HNR35 -0.53287118 -0.116395288 0.218316751 0.111462787 0.82810219
## SHR 0.35389648 -0.181537883 0.136893713 0.087188543 -0.58077659
## soe -0.40316847 -0.004677426 0.007849427 0.117966357 0.93424725
## Z1mnZ0 1.00000000 -0.479471463 0.132402681 0.143899368 -0.25634372
## Z2mnZ1 -0.47947146 1.000000000 -0.901393170 -0.176529973 -0.08020731
## Z3mnZ2 0.13240268 -0.901393170 1.000000000 -0.050618065 0.01898548
## Z4mnZ3 0.14389937 -0.176529973 -0.050618065 1.000000000 0.15981826
## F0Bark -0.25634372 -0.080207311 0.018985484 0.159818261 1.00000000
## A1mnA2 0.15440193 -0.588053094 0.710977937 -0.040573920 -0.50258069
## A1mnA3 -0.06129172 0.361831277 -0.361830045 0.355533137 -0.61872264
## A2mnA3 -0.17662983 0.769563482 -0.873375083 0.302455298 -0.04228227
## A1mnA2 A1mnA3 A2mnA3
## CPP -0.46294027 -0.35589200 0.122501972
## Energy 0.18810560 0.03344952 -0.133626988
## H1A1c -0.55266962 -0.66446955 -0.034489484
## H1A2c 0.80194637 -0.21090543 -0.836352608
## H1A3c -0.19894208 0.74253350 0.728126945
## H1H2c -0.29133499 -0.55795935 -0.174847909
## H2H4c 0.16802747 0.30498923 0.088160327
## H2KH5Kc 0.68460673 0.03925946 -0.548549664
## H42Kc -0.44401978 0.44524464 0.710845732
## HNR05 -0.50509144 -0.58870128 -0.017515845
## HNR15 -0.51038750 -0.34659555 0.169584736
## HNR25 -0.32063266 -0.47484763 -0.087411661
## HNR35 -0.23532567 -0.47720189 -0.161231042
## SHR 0.45272098 0.42180815 -0.064148964
## soe -0.46030423 -0.52272126 -0.005568759
## Z1mnZ0 0.15440193 -0.06129172 -0.176629833
## Z2mnZ1 -0.58805309 0.36183128 0.769563482
## Z3mnZ2 0.71097794 -0.36183005 -0.873375083
## Z4mnZ3 -0.04057392 0.35553314 0.302455298
## F0Bark -0.50258069 -0.61872264 -0.042282270
## A1mnA2 1.00000000 0.22151418 -0.677425247
## A1mnA3 0.22151418 1.00000000 0.567258129
## A2mnA3 -0.67742525 0.56725813 1.000000000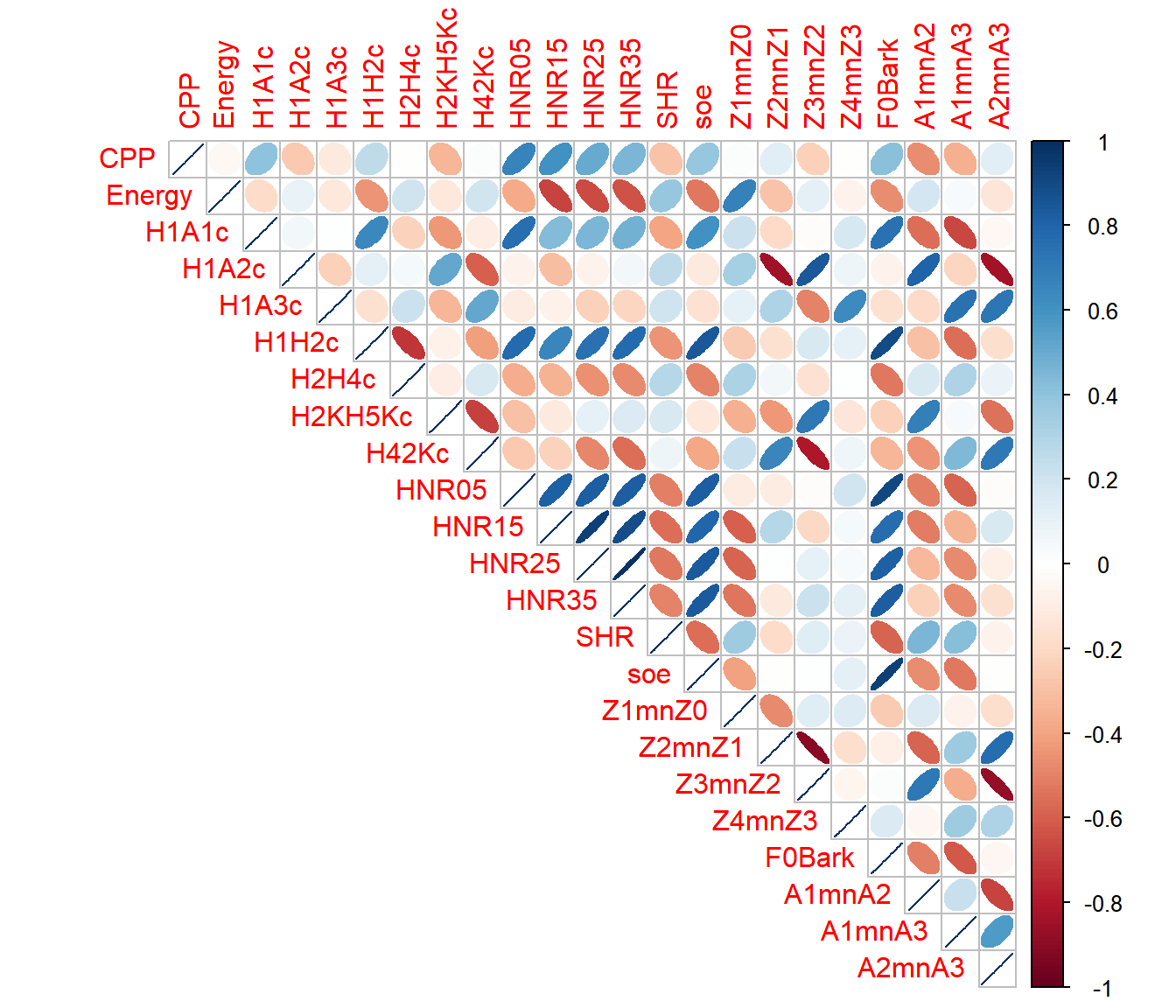
9.3.1.2 Correlation plots 2
Let’s first compute the correlations between all numeric variables and plot these with the p values
### correlation using "corrplot"
### based on the function `rcorr' from the `Hmisc` package
### Need to change dataframe into a matrix
corr <-
dfPharV2 %>%
select(where(is.numeric)) %>%
data.matrix(dfPharV2) %>%
rcorr(type = "pearson")
print(corr)## CPP Energy H1A1c H1A2c H1A3c H1H2c H2H4c H2KH5Kc H42Kc HNR05 HNR15
## CPP 1.00 -0.03 0.40 -0.26 -0.11 0.25 -0.01 -0.34 0.02 0.67 0.60
## Energy -0.03 1.00 -0.19 0.09 -0.12 -0.44 0.20 -0.13 0.19 -0.37 -0.67
## H1A1c 0.40 -0.19 1.00 0.05 0.01 0.65 -0.22 -0.43 -0.09 0.76 0.43
## H1A2c -0.26 0.09 0.05 1.00 -0.23 0.12 0.04 0.51 -0.60 -0.06 -0.30
## H1A3c -0.11 -0.12 0.01 -0.23 1.00 -0.16 0.21 -0.33 0.51 -0.11 -0.08
## H1H2c 0.25 -0.44 0.65 0.12 -0.16 1.00 -0.71 -0.08 -0.42 0.77 0.65
## H2H4c -0.01 0.20 -0.22 0.04 0.21 -0.71 1.00 -0.10 0.16 -0.37 -0.34
## H2KH5Kc -0.34 -0.13 -0.43 0.51 -0.33 -0.08 -0.10 1.00 -0.68 -0.30 -0.12
## H42Kc 0.02 0.19 -0.09 -0.60 0.51 -0.42 0.16 -0.68 1.00 -0.26 -0.23
## HNR05 0.67 -0.37 0.76 -0.06 -0.11 0.77 -0.37 -0.30 -0.26 1.00 0.81
## HNR15 0.60 -0.67 0.43 -0.30 -0.08 0.65 -0.34 -0.12 -0.23 0.81 1.00
## HNR25 0.50 -0.65 0.45 -0.06 -0.23 0.75 -0.45 0.11 -0.48 0.82 0.94
## HNR35 0.45 -0.63 0.47 0.06 -0.22 0.78 -0.47 0.16 -0.55 0.82 0.89
## SHR -0.28 0.38 -0.40 0.26 0.21 -0.44 0.28 0.17 0.08 -0.50 -0.56
## soe 0.39 -0.52 0.61 -0.11 -0.15 0.84 -0.50 -0.13 -0.38 0.83 0.79
## Z1mnZ0 0.01 0.67 0.21 0.34 0.11 -0.26 0.31 -0.35 0.23 -0.10 -0.60
## Z2mnZ1 0.12 -0.28 -0.20 -0.85 0.31 -0.17 0.05 -0.44 0.65 -0.11 0.28
## Z3mnZ2 -0.24 0.12 -0.01 0.84 -0.50 0.16 -0.15 0.72 -0.80 -0.01 -0.21
## Z4mnZ3 -0.01 -0.06 0.17 0.08 0.63 0.11 0.00 -0.13 0.06 0.19 0.04
## F0Bark 0.42 -0.46 0.74 -0.07 -0.16 0.89 -0.52 -0.24 -0.33 0.90 0.76
## A1mnA2 -0.46 0.19 -0.55 0.80 -0.20 -0.29 0.17 0.68 -0.44 -0.51 -0.51
## A1mnA3 -0.36 0.03 -0.66 -0.21 0.74 -0.56 0.30 0.04 0.45 -0.59 -0.35
## A2mnA3 0.12 -0.13 -0.03 -0.84 0.73 -0.17 0.09 -0.55 0.71 -0.02 0.17
## HNR25 HNR35 SHR soe Z1mnZ0 Z2mnZ1 Z3mnZ2 Z4mnZ3 F0Bark A1mnA2
## CPP 0.50 0.45 -0.28 0.39 0.01 0.12 -0.24 -0.01 0.42 -0.46
## Energy -0.65 -0.63 0.38 -0.52 0.67 -0.28 0.12 -0.06 -0.46 0.19
## H1A1c 0.45 0.47 -0.40 0.61 0.21 -0.20 -0.01 0.17 0.74 -0.55
## H1A2c -0.06 0.06 0.26 -0.11 0.34 -0.85 0.84 0.08 -0.07 0.80
## H1A3c -0.23 -0.22 0.21 -0.15 0.11 0.31 -0.50 0.63 -0.16 -0.20
## H1H2c 0.75 0.78 -0.44 0.84 -0.26 -0.17 0.16 0.11 0.89 -0.29
## H2H4c -0.45 -0.47 0.28 -0.50 0.31 0.05 -0.15 0.00 -0.52 0.17
## H2KH5Kc 0.11 0.16 0.17 -0.13 -0.35 -0.44 0.72 -0.13 -0.24 0.68
## H42Kc -0.48 -0.55 0.08 -0.38 0.23 0.65 -0.80 0.06 -0.33 -0.44
## HNR05 0.82 0.82 -0.50 0.83 -0.10 -0.11 -0.01 0.19 0.90 -0.51
## HNR15 0.94 0.89 -0.56 0.79 -0.60 0.28 -0.21 0.04 0.76 -0.51
## HNR25 1.00 0.99 -0.53 0.84 -0.59 0.01 0.11 0.04 0.81 -0.32
## HNR35 0.99 1.00 -0.50 0.84 -0.53 -0.12 0.22 0.11 0.83 -0.24
## SHR -0.53 -0.50 1.00 -0.56 0.35 -0.18 0.14 0.09 -0.58 0.45
## soe 0.84 0.84 -0.56 1.00 -0.40 0.00 0.01 0.12 0.93 -0.46
## Z1mnZ0 -0.59 -0.53 0.35 -0.40 1.00 -0.48 0.13 0.14 -0.26 0.15
## Z2mnZ1 0.01 -0.12 -0.18 0.00 -0.48 1.00 -0.90 -0.18 -0.08 -0.59
## Z3mnZ2 0.11 0.22 0.14 0.01 0.13 -0.90 1.00 -0.05 0.02 0.71
## Z4mnZ3 0.04 0.11 0.09 0.12 0.14 -0.18 -0.05 1.00 0.16 -0.04
## F0Bark 0.81 0.83 -0.58 0.93 -0.26 -0.08 0.02 0.16 1.00 -0.50
## A1mnA2 -0.32 -0.24 0.45 -0.46 0.15 -0.59 0.71 -0.04 -0.50 1.00
## A1mnA3 -0.47 -0.48 0.42 -0.52 -0.06 0.36 -0.36 0.36 -0.62 0.22
## A2mnA3 -0.09 -0.16 -0.06 -0.01 -0.18 0.77 -0.87 0.30 -0.04 -0.68
## A1mnA3 A2mnA3
## CPP -0.36 0.12
## Energy 0.03 -0.13
## H1A1c -0.66 -0.03
## H1A2c -0.21 -0.84
## H1A3c 0.74 0.73
## H1H2c -0.56 -0.17
## H2H4c 0.30 0.09
## H2KH5Kc 0.04 -0.55
## H42Kc 0.45 0.71
## HNR05 -0.59 -0.02
## HNR15 -0.35 0.17
## HNR25 -0.47 -0.09
## HNR35 -0.48 -0.16
## SHR 0.42 -0.06
## soe -0.52 -0.01
## Z1mnZ0 -0.06 -0.18
## Z2mnZ1 0.36 0.77
## Z3mnZ2 -0.36 -0.87
## Z4mnZ3 0.36 0.30
## F0Bark -0.62 -0.04
## A1mnA2 0.22 -0.68
## A1mnA3 1.00 0.57
## A2mnA3 0.57 1.00
##
## n= 402
##
##
## P
## CPP Energy H1A1c H1A2c H1A3c H1H2c H2H4c H2KH5Kc H42Kc HNR05
## CPP 0.4968 0.0000 0.0000 0.0229 0.0000 0.9131 0.0000 0.7007 0.0000
## Energy 0.4968 0.0002 0.0662 0.0141 0.0000 0.0000 0.0121 0.0000 0.0000
## H1A1c 0.0000 0.0002 0.2743 0.8861 0.0000 0.0000 0.0000 0.0695 0.0000
## H1A2c 0.0000 0.0662 0.2743 0.0000 0.0198 0.3906 0.0000 0.0000 0.2180
## H1A3c 0.0229 0.0141 0.8861 0.0000 0.0009 0.0000 0.0000 0.0000 0.0303
## H1H2c 0.0000 0.0000 0.0000 0.0198 0.0009 0.0000 0.1186 0.0000 0.0000
## H2H4c 0.9131 0.0000 0.0000 0.3906 0.0000 0.0000 0.0545 0.0011 0.0000
## H2KH5Kc 0.0000 0.0121 0.0000 0.0000 0.0000 0.1186 0.0545 0.0000 0.0000
## H42Kc 0.7007 0.0000 0.0695 0.0000 0.0000 0.0000 0.0011 0.0000 0.0000
## HNR05 0.0000 0.0000 0.0000 0.2180 0.0303 0.0000 0.0000 0.0000 0.0000
## HNR15 0.0000 0.0000 0.0000 0.0000 0.1308 0.0000 0.0000 0.0207 0.0000 0.0000
## HNR25 0.0000 0.0000 0.0000 0.2235 0.0000 0.0000 0.0000 0.0325 0.0000 0.0000
## HNR35 0.0000 0.0000 0.0000 0.2678 0.0000 0.0000 0.0000 0.0016 0.0000 0.0000
## SHR 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0006 0.1316 0.0000
## soe 0.0000 0.0000 0.0000 0.0215 0.0021 0.0000 0.0000 0.0106 0.0000 0.0000
## Z1mnZ0 0.7758 0.0000 0.0000 0.0000 0.0281 0.0000 0.0000 0.0000 0.0000 0.0371
## Z2mnZ1 0.0145 0.0000 0.0000 0.0000 0.0000 0.0007 0.2963 0.0000 0.0000 0.0291
## Z3mnZ2 0.0000 0.0199 0.7761 0.0000 0.0000 0.0011 0.0025 0.0000 0.0000 0.7981
## Z4mnZ3 0.8516 0.2106 0.0005 0.1306 0.0000 0.0277 0.9449 0.0089 0.2068 0.0000
## F0Bark 0.0000 0.0000 0.0000 0.1629 0.0011 0.0000 0.0000 0.0000 0.0000 0.0000
## A1mnA2 0.0000 0.0001 0.0000 0.0000 0.0000 0.0000 0.0007 0.0000 0.0000 0.0000
## A1mnA3 0.0000 0.5036 0.0000 0.0000 0.0000 0.0000 0.0000 0.4325 0.0000 0.0000
## A2mnA3 0.0140 0.0073 0.4905 0.0000 0.0000 0.0004 0.0775 0.0000 0.0000 0.7262
## HNR15 HNR25 HNR35 SHR soe Z1mnZ0 Z2mnZ1 Z3mnZ2 Z4mnZ3 F0Bark
## CPP 0.0000 0.0000 0.0000 0.0000 0.0000 0.7758 0.0145 0.0000 0.8516 0.0000
## Energy 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0199 0.2106 0.0000
## H1A1c 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.7761 0.0005 0.0000
## H1A2c 0.0000 0.2235 0.2678 0.0000 0.0215 0.0000 0.0000 0.0000 0.1306 0.1629
## H1A3c 0.1308 0.0000 0.0000 0.0000 0.0021 0.0281 0.0000 0.0000 0.0000 0.0011
## H1H2c 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0007 0.0011 0.0277 0.0000
## H2H4c 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2963 0.0025 0.9449 0.0000
## H2KH5Kc 0.0207 0.0325 0.0016 0.0006 0.0106 0.0000 0.0000 0.0000 0.0089 0.0000
## H42Kc 0.0000 0.0000 0.0000 0.1316 0.0000 0.0000 0.0000 0.0000 0.2068 0.0000
## HNR05 0.0000 0.0000 0.0000 0.0000 0.0000 0.0371 0.0291 0.7981 0.0000 0.0000
## HNR15 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3872 0.0000
## HNR25 0.0000 0.0000 0.0000 0.0000 0.0000 0.8436 0.0285 0.4726 0.0000
## HNR35 0.0000 0.0000 0.0000 0.0000 0.0000 0.0196 0.0000 0.0254 0.0000
## SHR 0.0000 0.0000 0.0000 0.0000 0.0000 0.0003 0.0060 0.0808 0.0000
## soe 0.0000 0.0000 0.0000 0.0000 0.0000 0.9255 0.8753 0.0180 0.0000
## Z1mnZ0 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0079 0.0038 0.0000
## Z2mnZ1 0.0000 0.8436 0.0196 0.0003 0.9255 0.0000 0.0000 0.0004 0.1083
## Z3mnZ2 0.0000 0.0285 0.0000 0.0060 0.8753 0.0079 0.0000 0.3114 0.7043
## Z4mnZ3 0.3872 0.4726 0.0254 0.0808 0.0180 0.0038 0.0004 0.3114 0.0013
## F0Bark 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1083 0.7043 0.0013
## A1mnA2 0.0000 0.0000 0.0000 0.0000 0.0000 0.0019 0.0000 0.0000 0.4172 0.0000
## A1mnA3 0.0000 0.0000 0.0000 0.0000 0.0000 0.2201 0.0000 0.0000 0.0000 0.0000
## A2mnA3 0.0006 0.0800 0.0012 0.1993 0.9114 0.0004 0.0000 0.0000 0.0000 0.3978
## A1mnA2 A1mnA3 A2mnA3
## CPP 0.0000 0.0000 0.0140
## Energy 0.0001 0.5036 0.0073
## H1A1c 0.0000 0.0000 0.4905
## H1A2c 0.0000 0.0000 0.0000
## H1A3c 0.0000 0.0000 0.0000
## H1H2c 0.0000 0.0000 0.0004
## H2H4c 0.0007 0.0000 0.0775
## H2KH5Kc 0.0000 0.4325 0.0000
## H42Kc 0.0000 0.0000 0.0000
## HNR05 0.0000 0.0000 0.7262
## HNR15 0.0000 0.0000 0.0006
## HNR25 0.0000 0.0000 0.0800
## HNR35 0.0000 0.0000 0.0012
## SHR 0.0000 0.0000 0.1993
## soe 0.0000 0.0000 0.9114
## Z1mnZ0 0.0019 0.2201 0.0004
## Z2mnZ1 0.0000 0.0000 0.0000
## Z3mnZ2 0.0000 0.0000 0.0000
## Z4mnZ3 0.4172 0.0000 0.0000
## F0Bark 0.0000 0.0000 0.3978
## A1mnA2 0.0000 0.0000
## A1mnA3 0.0000 0.0000
## A2mnA3 0.0000 0.0000## use corrplot to obtain a nice correlation plot!
corrplot(corr$r, p.mat = corr$P,
addCoef.col = "black", diag = FALSE, type = "upper", tl.srt = 55)